
Recently, the Bioinformatics Center team of Jinfeng Laboratory and the scientific research team of Sun Yat-sen University published a research paper titled "SpaBalance: Balanced Learning for Efficient Spatial Multi-Omics Decoding" in the international journal Advanced Science, proposing a new AI computing framework for efficient integration of spatial multi-omics data, which is expected to significantly improve the identification and understanding of spatial structures and molecular regulatory networks in tissue samples.
In recent years, with the maturity of spatial transcriptomics, spatial proteomics, spatial epigenomics and other technologies, researchers can simultaneously obtain different omics layers in tissues (such as gene expression, protein levels, chromatin status, etc.) and their spatial distribution information. This "multi-layer + spatial" data provides an unprecedented perspective on cell type distribution, tissue microenvironment regulation, development/disease processes, etc. However, there are great challenges in integrating these heterogeneous omics data and maintaining their spatial consistency. There are huge differences in data dimensions, distribution, and signal intensity between different omics. Traditional integration methods are often difficult to take into account shared and specific signals, and it is easy for a certain omics to "dominate" the final result, thereby masking important information of other modalities.
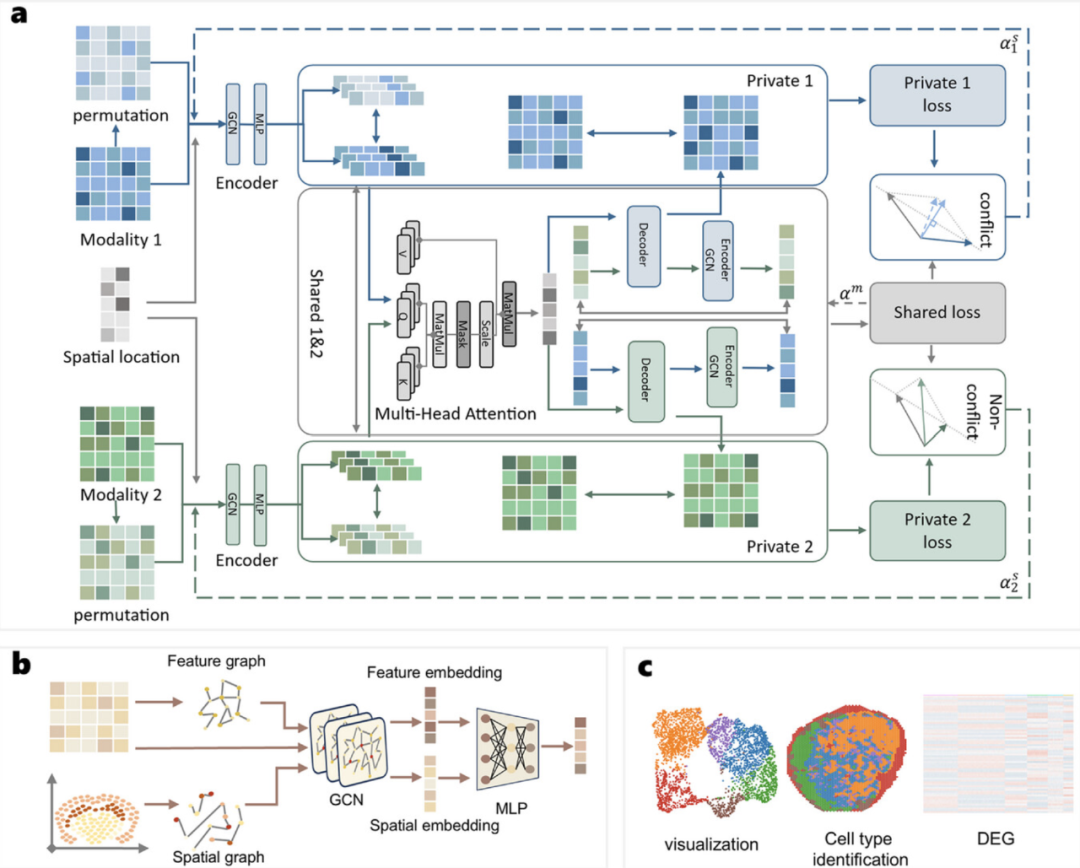
To solve the above problems, the research team developed SpaBalance, a unified integration framework for spatial multi-omics data. Its main innovations include:
• Gradient equilibrium mechanism: During the training process, SpaBalance dynamically adjusts the learning contributions between different omics. It does not rely on manual weight setting, but automatically "balances" based on the gradient information of different omics, effectively resolving conflicts and ensuring that each omics can be fully learned.
• Dual-stream feature decomposition structure: SpaBalance learns "shared representation across omics" and "omics-specific features" in parallel, which can not only capture the commonalities between different omics, but also retain their unique signals.
• Spatial graph + multi-head attention mechanism: combine spatial coordinates and omics features to construct a spatial graph, and integrate cross-omics information through multi-head attention to take into account spatial neighborhood relationships and complementary molecular features.
Through this design, SpaBalance transforms heterogeneous omics data into a unified and biologically meaningful low-dimensional representation (embedding), which can be used for downstream analysis such as spatial structure partitioning, cell type identification, and differential expression analysis.
"Advanced Science" is a well-known open source journal under Wiley Publishing House (impact factor 14.1, JCR Q1 area). It is committed to publishing top-level basic and applied research results in the fields of materials science, physics, chemistry, medicine, life sciences and engineering, and has high international academic influence.
Click on the lower left to read the original text and access the original text link.
https://advanced.onlinelibrary.wiley.com/doi/10.1002/advs.202512973